Download: https://osdyn.ifremer.fr/pyweb/notebooks/utils/plot/colorbar.ipynb
colorbars
How to specify the levels of a colorbar ?
[1]:
import matplotlib as mpl
import matplotlib.pyplot as plt
from osdyn.config import data_sample
from osdyn.grcm.generic import moawi
from osdyn.utils.plot.colorbar import auto_scale, histo
from osdyn.utils.plot.colormap import get_cmap
[2]:
mc = moawi(data_sample("mars_siggen.nc"), model="mars", varnames=["temp_sfc", "temp_bot"])
var = mc.ds.temp_bot
Found 1 files
[WARNING]: cf.py in name2key - line: 1288
No osdyn key for this variable : sc_w
[WARNING]: cf.py in name2key - line: 1288
No osdyn key for this variable : sc_t
[3]:
mc.ds
[3]:
<xarray.Dataset>
Dimensions: (nz_g: 41, nz_c: 40, nt: 3, ny_c: 33, nx_c: 33, nx_g: 33, ny_g: 33)
Coordinates:
* nz_g (nz_g) float64 0.5 1.5 2.5 3.5 4.5 ... 36.5 37.5 38.5 39.5 40.5
* nz_c (nz_c) float64 1.0 2.0 3.0 4.0 5.0 ... 36.0 37.0 38.0 39.0 40.0
* nt (nt) int64 0 1 2
lat_t (ny_c, nx_c) float64 47.64 47.64 47.64 47.64 ... 49.08 49.08 49.08
lon_t (ny_c, nx_c) float64 -6.249 -6.182 -6.115 ... -4.237 -4.17 -4.102
* nx_c (nx_c) float32 1.0 11.0 21.0 31.0 41.0 ... 291.0 301.0 311.0 321.0
* ny_c (ny_c) float32 1.0 11.0 21.0 31.0 41.0 ... 291.0 301.0 311.0 321.0
time (nt) float64 3.524e+09 3.524e+09 3.524e+09
* nx_g (nx_g) float32 1.5 11.5 21.5 31.5 41.5 ... 291.5 301.5 311.5 321.5
* ny_g (ny_g) float32 1.5 11.5 21.5 31.5 41.5 ... 291.5 301.5 311.5 321.5
sc_t (nz_c) float32 -0.9875 -0.9625 -0.9375 ... -0.0625 -0.0375 -0.0125
sc_w (nz_g) float32 -1.0 -0.975 -0.95 -0.925 ... -0.05 -0.025 0.0
Data variables:
h_t (ny_c, nx_c) float32 -152.0 -159.6 -150.8 ... -95.97 -90.55 -92.11
theta float64 6.0
b float64 0.0
sc_cs_t (nz_c) float32 -0.9277 -0.7985 -0.6873 ... -0.001125 -0.0003722
sc_cs_w (nz_g) float32 -1.0 -0.8607 -0.7408 ... -0.00151 -0.0007464 0.0
hc (ny_c, nx_c) float32 9.0 9.0 9.0 9.0 9.0 ... 9.0 9.0 9.0 9.0 9.0
temp_sfc (nt, ny_c, nx_c) float32 nan nan nan nan ... 14.56 14.67 14.72
temp_bot (nt, ny_c, nx_c) float32 nan nan nan nan ... 14.54 14.63 14.71
Attributes:
model: MarsClassic quick search
The data are used to get the minimum and maximum values
[4]:
scale = auto_scale(var)
scale
[4]:
array([11., 12., 13., 14., 15., 16., 17., 18.])
[5]:
def plot(scale):
cmap = get_cmap("cmo.thermal")
norm = mpl.colors.BoundaryNorm(scale, ncolors=cmap.N, clip=True)
plt.figure()
c = plt.imshow(var[-1,...], cmap="cmo.thermal", norm=norm)
# plt.contour(mc.ds.temp_sfc.values[-1,...], cmap='cmo.thermal', levels=scale)
plt.colorbar(c)
plt.show()
[6]:
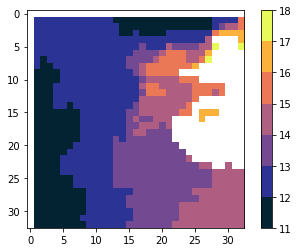
plot(scale)
[7]:
scale = auto_scale(var, nmax=11)
scale
[7]:
array([11., 12., 13., 14., 15., 16., 17., 18.])
[8]:
scale_n19 = auto_scale(var, nmax=19)
scale_n19
[8]:
array([11. , 11.5, 12. , 12.5, 13. , 13.5, 14. , 14.5, 15. , 15.5, 16. ,
16.5, 17. , 17.5])
[9]:
plot(scale_n19)
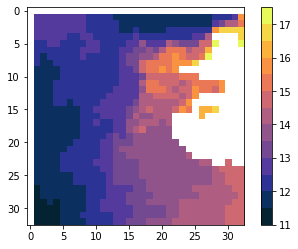
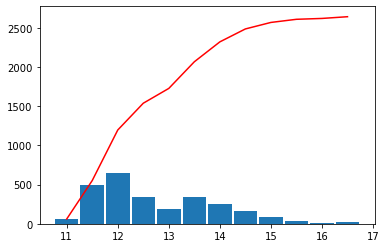
The colorbar separators flatten the histogram of the data
Get the range
[10]:
scalehist = auto_scale(var, nmax=12, flathisto=True, **{"show": False})
scalehist
[10]:
array([11., 12., 13., 14., 15., 16., 17.])
[11]:
scalehist = auto_scale(var, nmax=20, flathisto=True, **{"show": True})
scalehist
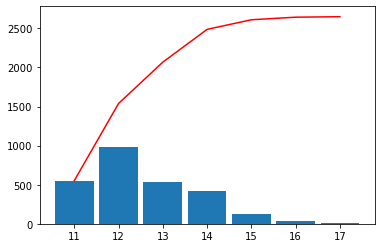
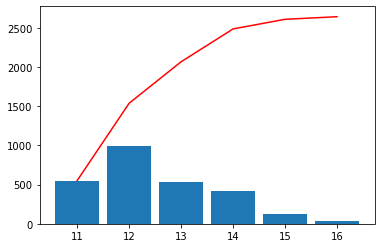


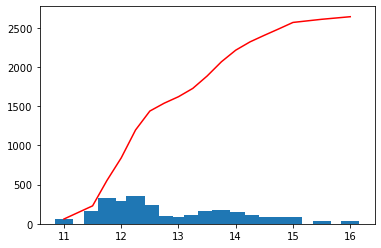
[11]:
array([11. , 11.5 , 11.75, 12. , 12.25, 12.5 , 12.75, 13. , 13.25,
13.5 , 13.75, 14. , 14.25, 14.5 , 14.75, 15. , 15.5 , 16. ,
17. ])
[12]:
len(scalehist)
[12]:
19
[13]:
plot(scalehist)
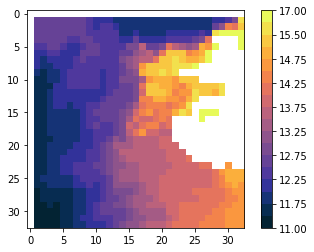
The scale without flattening the histogram differs although we ask for the same number of separators and the same bounds
[14]:
scale_n19 = auto_scale(var, nmax=19)
scale_n19
[14]:
array([11. , 11.5, 12. , 12.5, 13. , 13.5, 14. , 14.5, 15. , 15.5, 16. ,
16.5, 17. , 17.5])
[15]:
scale_n19 = auto_scale(var, nmax=19, vmin=11, vmax=17)
scale_n19
[15]:
array([11. , 11.5, 12. , 12.5, 13. , 13.5, 14. , 14.5, 15. , 15.5, 16. ,
16.5, 17. ])
Check the repartition of the values
[16]:
hist, bins = histo(var, bins=scalehist, show=True)
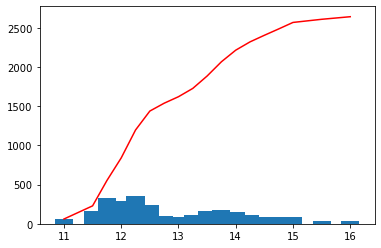
[17]:
hist, bins = histo(var, bins=scale_n19, show=True)